Concepts¶
One thing to bind them all¶
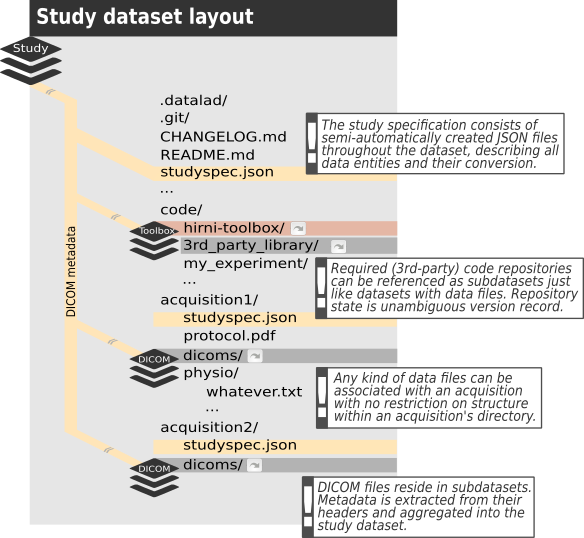
The central piece of the envisioned structure is a Datalad dataset, containing (i.e. referencing) all the raw data of a study alongside code, containers, protocols, metadata, … you name it. Everything that is making up your study and can be considered raw, in that it cannot be derived from other raw information that is already included. Furthermore, this dataset is supposed to have a subdirectory per each acquisition, which in turn should contain a subdataset for the DICOM files. All other data acquired besides (like physiological data, stimulation log files, etc.) as well as any other information specific to that acquisition should go beneath the respective acquisition subdirectory. Apart from that, it’s entirely up to you to decide about the structure within those acquisition directories.
This raw dataset will then be referenced by the converted (i.e. to BIDS) dataset the same way itself references the DICOM datasets. It’s recommended to follow this principle for further processing, analyses, etc. Let the results of a process be captured in a dataset and reference the input data as a subdataset. In that regard think of datasets as software packages and subdatasets as dependencies. In other words: Learn from YODA!
The second piece is the study specification. This is an additional metadata layer, the conversion of the dataset will be based on. It consists of JSON files within the dataset and describes contained data (files, directories or “logical entities”) in terms of the conversion target. For example in case of BIDS as the targeted layout it should assign the BIDS terms used by BIDS to determine file names and locations to each image series contained in the DICOM files. In addition the specification allows to define (custom) conversion routines for each data entity it describes. Furthermore, it can carry any additional metadata you might have no other place to record (like a comment about an aborted scan) or that you might need to be available to your conversion routine for a particular type of file.
Finally, by default a toolbox dataset will be part of the study dataset, providing a collection of conversion routines and container images to run them in, which can be referenced from within the study specification. This isn’t mandatory, though, but rather a default for convenience. You can have your own toolbox or none at all.
Trust automation¶
Binding everything in that one dataset will allow us to use automatically generated references to particular versions of each file for provenance capture. This is mostly relying on DataLad’s run command, which runs arbitrary executables and produces a (machine readable) record of the exact version of inputs, the executed call, possibly the container it ran in and its results. Thereby you can trace back the provenance of all derived data files throughout the entire history of the dataset (and its subdatasets) and reproduce the results from scratch. Another aspect of automation is the creation of the above mentioned study specification. Datalad-hirni comes with a routine to import DICOM files, extract the header data as Datalad metadata and derive a specification for the contained image series for conversion to BIDS.
Conversion¶
The conversion of a study dataset was build with BIDS in mind, but technically the mechanism can target any other layout specification (or another version of the same standard). The idea here is to create a new dataset, which is to become the converted dataset. The original study dataset is then linked into it as a subdataset. So, again the result of the conversion process is referencing the exact version of the data it is based upon. The conversion is executing whatever is specified in the study dataset’s specification and is executed per specification file, allowing to convert a single acquisition or even just a single file only, for example. In addition an anonymization can be included in the conversion.
Don’t trust automation¶
While automation can help a lot, in reality there’s no “one size fits it all”. Rules to derive BIDS terms from DICOM headers won’t capture all cases, information no one included at the scanner can not be made up by an automation routine and there’s a lot of potential for human error. This is why many adjustments can be made. First off, that is the reason for the specification to be added as a separate layer. If the automatically derived information is wrong, incomplete or insufficient, you can review that specification and edit or enhance it before even trying to convert anything. Since the specification is stored in JSON format, it can easily be edited programmatically as well as manually. To ease the manual review or edition Datalad-hirni comes with a browser based GUI for this purpose. Secondly, the rules to derive the specification from DICOM metadata are customizable. You can have your own rules or configure a set of such rules to be applied in a particular order. Also the specification of conversion routines is quite flexible. You can plug in whatever routine you like, the conversion can be an entire list of routines to execute, those routines have access to all the fields in the specification they are running on and their execution can depend on whether or not the anonymization switch was used for the conversion command. That way the default defacing - which also can be replaced by another routine, of course - is implemented, for example.
For a more detailed explanation of how it works and how to approach customization, you might be interested in watching this talk.